Project Grant 2016
New in vivo adapted approaches to reveal molecular mechanisms of bacterial persistence
Principal investigator:
Maria Fällman professor in microbiology
Co-investigators:
Karolinska Institutet
Mikael Rhen
Uppsala University
Mikael Sellin
Institution:
Umeå University
Grant in SEK:
SEK 28.7 million over five years
Maria Fällman, Professor of Medical Microbiology at Umeå University, has already shown how pathogenic bacteria can enter a host cell and resist its immune system. She has also identified host cell proteins that are key targets for infection-carrying bacteria.
“I’ve worked for a long time on fundamental questions like why micro-organisms make us ill,” Fällman says.
“Most of her experiments are carried out on the Yersinia pseudotuberculosis bacterium.
It is not an important spreader of disease; it is a model bacterium, used because it has been well studied, and particularly because there are good infection models. It also has roughly the same DNA as other types of pathogenic intestinal bacteria such as Salmonella, Shigella and E.coli.
“Yersinia has certain weapons that paralyze our immune cells. In our model system it invades intestinal tissue, and can survive there for up to four months. I want to know what enables the bacteria to live for so long,” Fällman explains.
Studies in living tissue
The grant that Fällman and her fellow researchers in Uppsala and Solna have been awarded by the Knut and Alice Wallenberg Foundation covers study of molecular mechanisms in Salmonella, which can be passed on from one person to another via water or food, and causes enormous harm to individuals and communities.
“We want to find the mechanisms that enable the bacteria to stay in the body and cause disease.”
Bacteria behave very differently in the test tube and in living tissue. The project will therefore be examining the two intestinal bacteria in a mouse model that involves processes similar to those in humans.
“Much of what we now know derives from laboratory experiments. As far as possible, we’ll be using methods that mimic a real infection. We hope our project will contribute a deeper understanding from experiments on living tissue. We’ll learn a lot of new things,” Fällman says.
The researchers will be infecting mice and isolating infected organs to map the genes used by the bacteria in that environment, and analyzing the mechanisms that predominate during persistent infections. They hope to find unique patterns of gene expression that differ from those found in the lab environment.
When they make a find, they will develop mutated bacteria that lack potentially important genes.
“We want to study how the mutated bacteria resist the stressful environment in real tissue where there is an active immune system. The gene products found to play a central role in infection of tissue may be potential target structures in developing new antimicrobial agents,” Fällman explains.
Agents of this kind will not kill the bacteria – merely stop them from remaining in the body. This will reduce the risk of resistance developing.
The project will also examine how the bacteria infect various organs, and study dispersal routes between the organs, as well as how these differ from one mutant to another.
Fluorescent bacteria
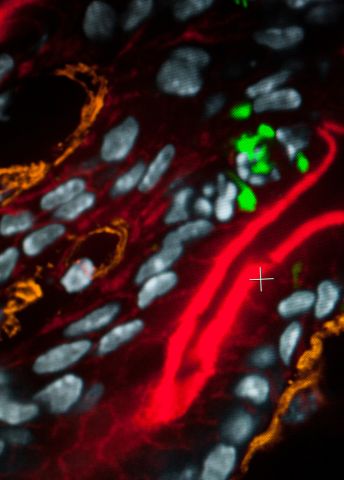
The researchers are using the new generation of sequencing to map the DNA and RNA of the bacteria. They will also be labeling gene products of interest with fluorescence, and examining them using advanced microscopy.
“If the bacteria are fluorescent, that means the gene is switched on. This will enable us to see the phases of an infection during which the gene is used. We hope it will give us fresh insights into mechanisms that are essential for bacteria to create an infection in tissue,” Fällman comments.
The project is still in its infancy, and will begin with small-scale pilot experiments.
“We’re feeling our way. No-one has done anything like this before, and we have prepared alternative plans in case we find something isn’t going as we expected. If it doesn’t work one way, maybe it’ll work another,” Fällman adds.
Text Carin Mannberg-Zackari
Translation Maxwell Arding
Photo Magnus Bergström
FACTS
The intestinal bacteria Yersinia pseudotuberculosis and Salmonella enterica Typhimurium are being studied to examine the ability of bacteria to infect and resist the immune system in living tissue.
Yersinia is a model bacterium, whose basic molecular mechanisms are fairly well studied.
Salmonellosis is a stomach disease, caused by various strains of salmonella bacteria. Resistance of stomach bacteria to antibiotics has become increasingly common in recent years. There is a pressing need for new drugs to combat bacterial infections.




