Strategic Grants
Human Protein Atlas Project
Proteins are important building blocks in human cells and are involved in virtually all bodily functions in both healthy and sick individuals. If they are malformed, or if their function is knocked out, they will be unable to fulfill their important role, which leads to various disorders.
The project is directed by Mathias Uhlén at KTH Royal Institute of Technology in Stockholm. Some 70 researchers are involved, primarily from KTH and Uppsala University.
Grant in SEK:
SEK 950 million
SEK 120 million in addition for 2025-2032
In total SEK 1.07 billion
Sweden has a proud tradition of conducting surveys that are fundamental to science. In the 18th century, Carl Linnaeus systematized all living creatures, plants and animals. He divided them into orders, genera and species. In the 19th century, the chemists took over. With Jöns Jacob Berzelius on the forefront, they looked for new elements. A total of 19 of the periodic system's 111 elements were found by Swedish researchers.
A gigantic project that will become historic is now being run from KTH Royal Institute of Technology in Stockholm and the Science for Life Laboratory (SciLifeLab) in Solna and Uppsala. In what is known as the Human Protein Atlas Project, the researchers are mapping every cog in the human machinery, all of the proteins that build and control our bodies.
“As of December 2013, we will have information on 80% of all human proteins. This is a milestone. The complete mapping will be finished at the end of 2015,” says Mathias Uhlén, Professor of Microbiology at KTH and head of the project that has been under way since 2003.
Proteins are the tools of life
In the 1990s, he was involved in the mapping of the human genome in the “Human Genome Project”. Through this project, researchers identified all of mankind's genes, some 20,000 genes in total. However, this is only the blueprint for what a human being should look like. Every gene contains a code for how amino acids should be joined together into a protein. It is then these proteins that carry out the gene's message. Some proteins function as building blocks, such as myosin and actin in the muscles. Other proteins, called enzymes, catalyze chemical reactions. Yet others work, for example, as hormones or signal substances, or sit in our senses and read odors, tastes and smells.
To fully understand how the human body works, we need to know how all of these proteins cooperate. In the 1990s, Mathias Uhlén began to wonder if it would be possible to map the human proteins in the same way that all genes had been mapped.
“We began to do some pilot studies and in 2002, we presented our ideas to the Knut and Alice Wallenberg Foundation,” says Mathias Uhlén.
They received initial financing and now, 10 years later, four-fifths of all proteins have been mapped. Half of them are new knowledge for the researchers.
“It is incredible. They are the same proteins that our ancestors had in their bodies and they are the proteins that our children will get,” says Mathias Uhlén.
Worldwide cooperation
Even if the project is run from Sweden, many countries are involved. The entire process begins with researchers at KTH inserting parts of a human gene in the genetic material of a bacterium; this gene has most often never been studied. The bacterium is then converted into a protein factory. With the gene as a blueprint, it produces the unknown protein. This is then sent off to South Korea or China. There, rabbits are vaccinated with the protein. The rabbits then produce antibodies that specifically recognize and attach to the unknown human protein.
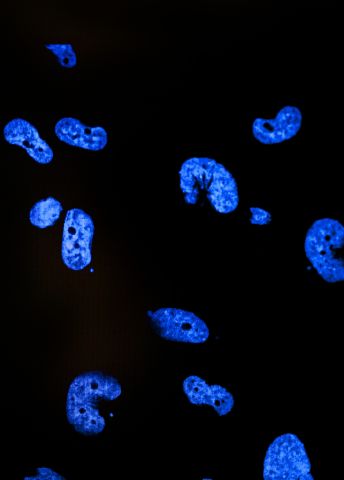
These antibodies are keys in the project. The researchers use them as a kind of molecular fishing rod. They connect fluorescent molecules to the antibodies, so that they can be traced, and then they go fishing for the unknown protein in tissue samples. Is the protein in the brain? In the heart? Or perhaps in the kidneys? And where in the cells is the protein? In the nucleus? On the surface of the cell? Or just a little everywhere?
The actual searching is done in three places in Sweden, Solna, Uppsala and Lund. Thanks to the fluorescence from the antibodies, the researchers can take pictures of where in the tissues various proteins are.
“So far, we have collected 13 million pictures. Every one of these has been manually reviewed by a pathologist in India,” says Mathias Uhlén.
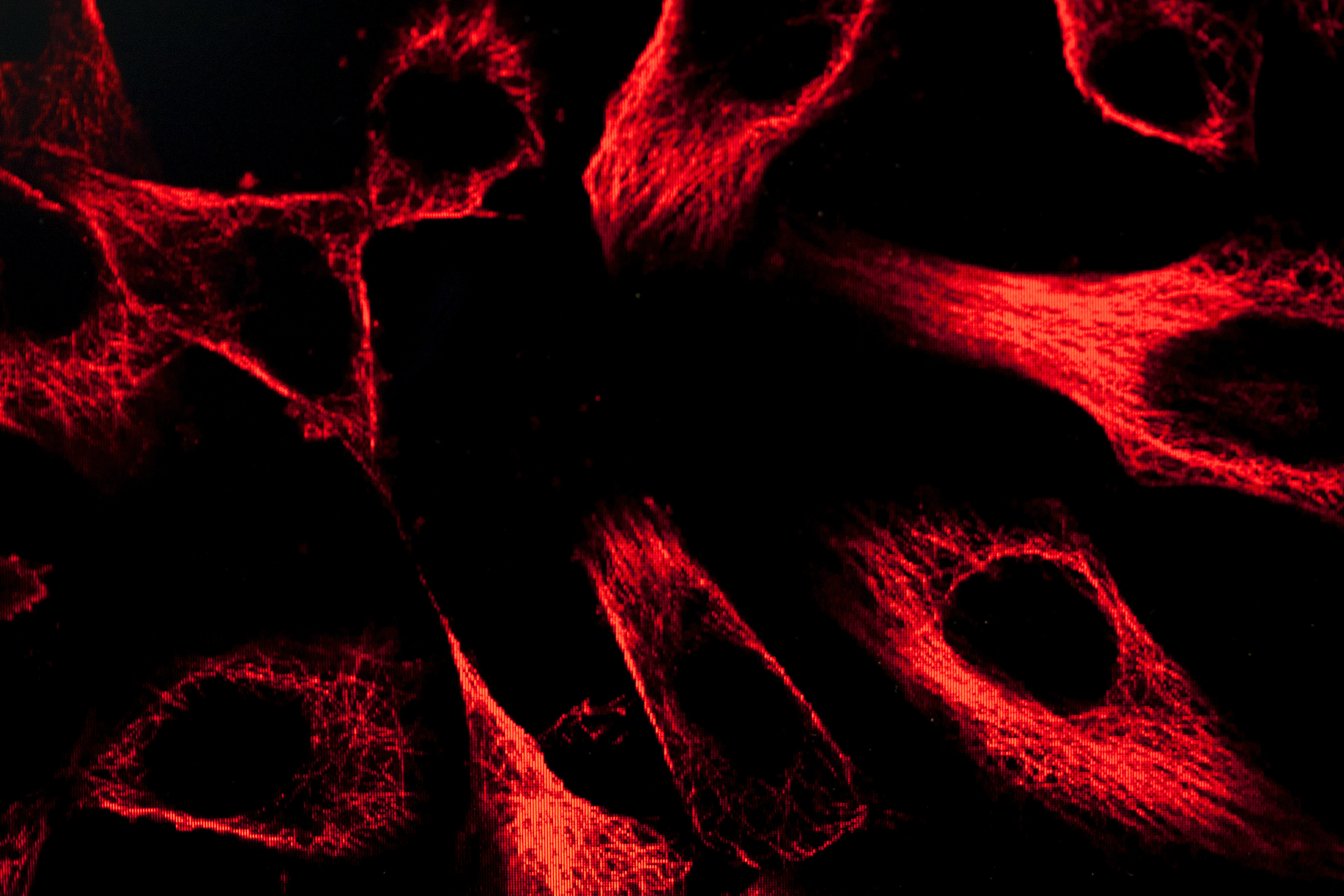
A database with both healthy and sick tissue
All information on various proteins is compiled and saved in an open database, www.proteinatlas.org, where anyone who wants to can study the information. In addition, researchers who themselves need to fish for something in a sample can purchase antibodies from the project via the company Atlas Antibodies AB:
“We send off around 100 antibodies a day and around once every two days, there is a publication in scientific journals that is based on antibodies from here,” says Mathias Uhlén.
The Human Protein Atlas Project has also been supplemented with a number of side projects. Among these, researchers are looking for proteins that can be linked to various kinds of cancer, Alzheimer's, rheumatoid arthritis, multiple sclerosis and a number of other diseases. The researchers use these discoveries for future studies that can lead to new knowledge about how the building-blocks in mankind's machinery work together. When researchers know which piece fails, they can more easily find methods to lubricate the machinery so that it begins to turn painlessly.
Text Ann Fernholm
Translation Semantix
Photo Magnus Bergström
More about Mathias Uhlén's research
Protein atlas maps the way to new drugs




