Project Grant 2020
Decoding bacterial toxin-antitoxin systems: from high-throughput discovery to molecular mechanisms and biotechnology
Principal investigator:
Dr. Gemma Atkinson
Co-investigators:
Lund University
Vasili Hauryliuk
Herwig Schüler
Umeå University
Magnus Wolf-Watz
Institution:
Lund University
Grant in SEK:
SEK 29.6 million over five years
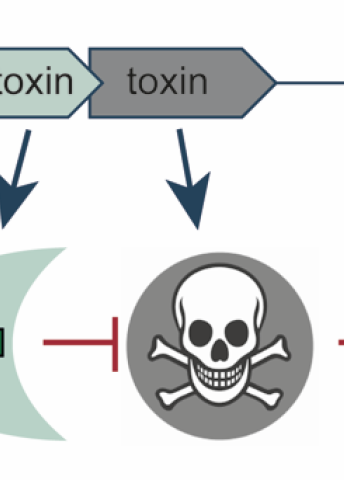
The existence of on-off switches in the DNA of many bacteria has long been known. These switches take the form of a gene pair, one of which codes for a toxin, the other for an antitoxin. The toxin acts as such, dramatically stopping bacterial growth. The antitoxin counters the effect by interacting with the toxin or by neutralizing its impact on the bacterial cell.
Much remains unknown about the diversity and function of different toxin-antitoxin gene pairs, and why bacteria have so many different varieties of them. They are hard to identify. No one knows how many there are.
Sifting through a mountain of data
Atkinson and her colleagues have received a grant from Knut and Alice Wallenberg Foundation to identify all variants of the toxin-antitoxin pair that they can detect, and ascertain their function in a cell. The computational tool she has developed is able to process large quantities of information, and identify new systems.
“Hundreds of thousands of genomes have been sequenced so far, but all kinds of secrets remain hidden from view. Advanced software is needed to find them.”
In addition to questions of how they work at the molecular level, another outstanding question is why these systems exists at all. Why does a bacterium want to use a toxin to poison itself?
“Bacteria don’t necessarily want to commit suicide – they use the toxin to halt a process. This may because the bacterium needs to conserve their resources when conditions are not optimal, or it may be to overcome a viral attack. When conditions improve, the bacterial population can continue growing with renewed vigor. The antitoxin is a very effective safety mechanism to keep the toxin in check when not needed.”
The Atkinson lab recently achieved a breakthrough in discovering a family of antitoxins that specialize in neutralizing hundreds or even thousands of unrelated toxins.
“We were really surprised. We have named this new antitoxin family Panacea after the Greek goddess of universal remedy. There are more mechanisms to be discovered. It’s exciting because you never know beforehand what you’re going to find.”
Much remains to be explored. How do the proteins work in detail – and are there more similar antitoxin families?
From new biology to new biotechnology
The project will add to our knowledge of the nature of bacteria, and the universe of protein functions that remain undiscovered. The result may be many new applications across a broad spectrum of fields.
“Controlling bacterial growth is important in biotech. There is also great medical potential. When antibiotics fail, we will need to create genetic weapons to kill pathogenic bacteria in patients. Phage therapy is an exciting prospect, and since toxin-antitoxin systems have a role in phage defense, we expect them to play a significant role in the future development of this technology”.
In his letter of support for the project application, Hans Adolfsson, vice-chancellor of Umeå University, expressed a hope that the project will have the same potential as the CRISPR-Cas9 gene editing tool. Professor Emmanuelle Charpentier was researching at Umeå University when she discovered and developed the system, for which she was awarded a joint Nobel Prize in Chemistry in 2020.
“The mechanisms are very different, but both processes can be used in biotech and medical applications. We believe we’ll find thousands of new toxin-antitoxin systems. That will give us an arsenal of tools for purposes such as controlling cell resources so the cell produces a protein or synthesizes a small molecule of interest to industry or in pharmaceutical manufacture.”
New phase in the project
This very day, Gemma Atkinson is looking forward to a meeting with a PhD student in her group who is developing an online web that enables other researchers to find toxin-antitoxin systems in their favorite bacterium. Both she and her husband Vasili Hauryliuk have had a busy summer moving their family and teams to Lund from Umeå, where they still have strong connections.
“The move to another environment is an exciting next step in our research. We are looking forward to all the new adventures and discoveries ahead.”
Text Carin Mannberg-Zackari
Translation Maxwell Arding
Photo Ola Nilsson, Vasili Hauryliuk, Gemma Atkinson
More about the project
Four teams at three institutions in two universities are collaborating on the study.
In the Department of Experimental Medical Sciences, Lund University, the Atkinson lab is taking on the bioinformatic and molecular evolutionary challenges, while Vasili Hauryliuk is driving the microbiology and biochemistry directions.
Toxin-antitoxin systems often contain proteins with unusual folds that have never been seen before, so structural biology is a significant part of the project. The structural biology directions in the project are being carried out by teams led by Magnus Wolf-Watz, in the Department of Chemistry, Umeå University and Herwig Schüler in the department of Biochemistry and Structural Biology, Lund University.