Project Grant 2017
Deciphering spatial signaling of protein clusters at the membrane
Principal investigator:
Björn Högberg, Professor of Molecular Systems Biophysics
Co-investigators:
Ana Teixeira
Simon Elsässer
Institution:
Karolinska Institutet
Grant in SEK:
SEK 25.2 million over five years
Cells can communicate with each other when a protein binds to a receptor on the cell membrane. This triggers a signal causing the cell to act in a given way.
But some signaling systems require that the signal not be sent until proteins have accumulated in large clusters around the receptor on the cell membrane – one or two proteins are not enough to activate the system. A research project is under way at Karolinska Institutet with the aim of developing methods to measure protein clusters of this kind.
“There are many areas where it is extremely useful to measure accumulation of proteins on the cell membrane, for example to understand how cancer cells communicate and how we can influence that communication,” says Björn Högberg, professor at the Department of Medical Biochemistry and Biophysics, Karolinska Institutet.
In breast cancer the prognosis is poorer when certain protein clusters form, and protein aggregations on a cell membrane in our immune system can render a cell harmless.
But Högberg points out that current methods of measuring these protein clusters are not really good enough. It is possible to used stained antibodies that bind to the proteins. But it is not then possible to see exactly how many proteins there are, or how they are placed. High-resolution microscopy is another option, but that requires advanced apparatus and complicated experiments.
“Our idea is to develop simpler methods that are statistically better and more objective than the present options.”
Imaging proteins without a microscope
The KI researchers’ methods are based on creating links between DNA strands that can then be deciphered using DNA sequencing. Högberg describes it as trying to study proteins at the molecular level with the help of sequencing.
"We try to get a DNA strand to form different sequences, depending on how the proteins are placed, so that we can read the DNA to obtain a picture of the proteins on the cell membrane.”
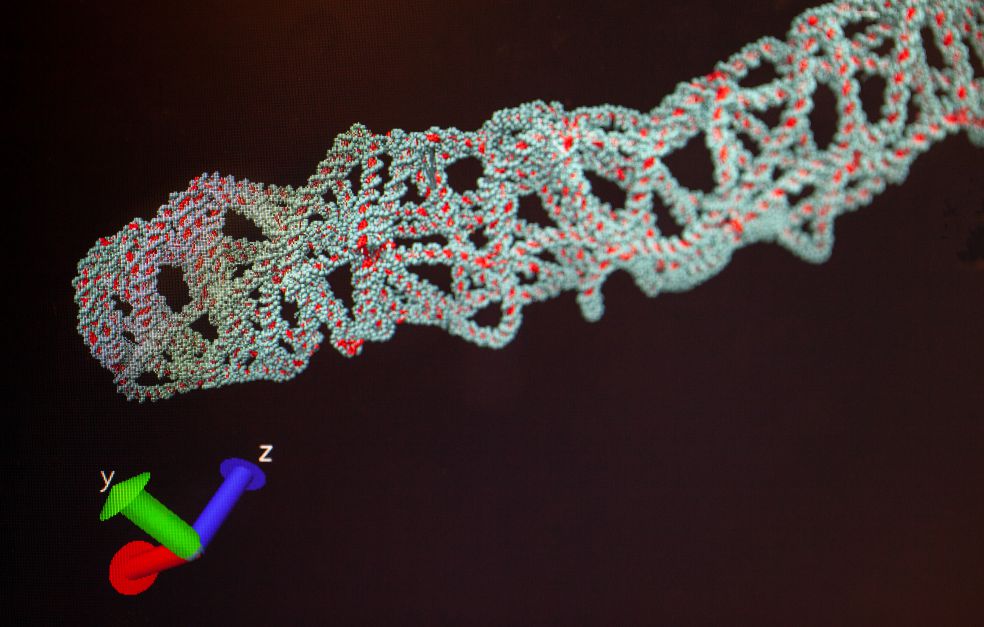
On Högberg’s desk lie what appear to be uncooked noodles. They are models of “DNA origami” – folded strands of DNA – a technique developed by his research team. DNA origami makes it easy for the researchers to build nanoscale structures, and design the exact form they should take.
DNA origami is used in one of the two methods that the researchers are now developing, primarily by the project’s co-investigator, Ana Texeira. The researchers take a flat origami structure, and attach short DNA strands that each contain a specific DNA sequence. The origami is then allowed to bind to the protein clusters the team wants to study, which have also been labeled with different DNA strands.
An enzyme – DNA polymerase – is then added. The enzyme can join up DNA strands, at the same time as the sample and the origami can contact one another. If the strands are close enough to each other, the information from the protein’s DNA will be copied to the DNA of the origami. Högberg explains:
“When we then remove the origami, we obtain a kind of imprint of the cell membrane written into the DNA of the origami. When we decipher the DNA using sequencing, we can essentially determine the molecular structure of the cell membrane – if DNA strand A is joined to strand B, then protein A and protein B must have been near one another, and so on.”
Easy to use
The other variant of the method does not use DNA origami as a carrier. But it is also based on labeling proteins with DNA strands, after which the polymerase enzyme joins the strands up so that sequencing of the new strand yields information about the placement of the proteins.
Högberg explains that this method can also be used to study molecules other than proteins. It provides an impression of how mRNA is arranged in cells, for example.
The methods might sound complicated, but in practice they are easy to use.
“You just take your tissue or cell samples, pipette some fluid onto them and sequence. The method’s simplicity is its strength. Using our methods we can easily study a large number of protein clusters at the same time. This yields statistically more reliable results than if you look at a selected cell under a microscope, plus it’s easy to carry out a large number of experiments in parallel.”
As an example, Högberg mentions the ability to swiftly test the effect hundreds of candidate drugs have on protein clusters in breast cancer cells.
Challenges in the lab
The project offers enormous potential, but also poses challenges. The researchers are taking on an entirely new concept – no one has yet published a study in which the viability of using sequencing to obtain data on molecular geometry is experimentally demonstrated.
“Members of our team have had to come up with a lot of their own ideas in the lab. There has been no one to teach us how to do it. We now see signs that the methods work, but we were taking a chance when we started out.”
In the cell culture lab researcher Ian Hoffecker places a culture dish under a microscope. So far, the researchers have found that their methods work in simple model systems. Before the project is over, Högberg expects them to have demonstrated that the methods also work on cell samples, and perhaps to have gone even further:
“It would be great to discover something new in the field of cancer biology as well.”
Text Sara Nilsson
Translation Maxwell Arding
Photo Magnus Bergström