Project Grant 2022
Learning the molecular component of the cell
Principal investigator:
Professor Arne Elofsson, Stockholm University
Co-investigators:
KTH Royal Institute of Technology
Hossein Azizpour
Lukas Käll
Uppsala University
Michael Landreh
Institution:
Stockholm University
Grant in SEK:
SEK 30 million over five years
The cell is the fundamental building block of life. It can be likened to a tiny, complex factory containing and producing thousands of molecular components. Proteins are the tools or workers in this “factory.” They perform numerous vital tasks, including building cell structures or carrying out chemical reactions.
Research into proteins has made strides in recent years. Scientists have managed to determine the structure of two hundred thousand or so individual proteins, along with many larger molecular complexes. However, more is needed to understand all cell functions fully.
Proteins display an enormous variety. The twenty thousand genes in the human genome generate innumerable protein forms through various kinds of modification. The proteins – consisting of just a few to several thousand amino acids – fold to form complex three-dimensional structures.
It still needs to be discovered precisely how many protein forms there are and which are essential. Researchers are keen to examine the complete variety of protein forms that may co-exist in a cell and also find out how they talk to each other – which is akin to understanding the molecular language of the cell.
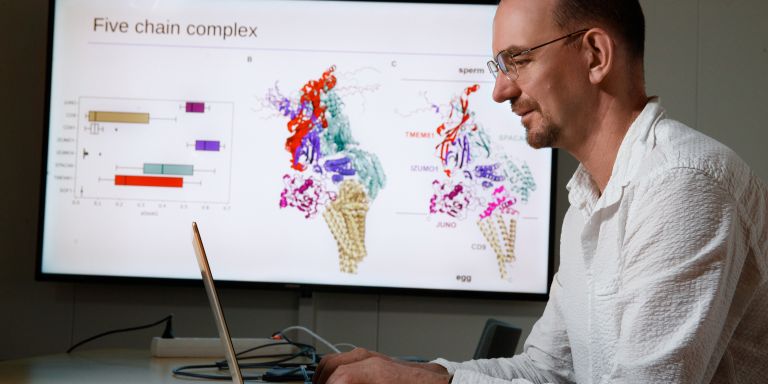
Arne Elofsson is a Professor of Bioinformatics at Stockholm University and is based at SciLifeLab, where he is leading a research project funded by Knut and Alice Wallenberg Foundation.
“We hope the project will successfully map most protein interactions occurring in a human cell. To achieve this, we will need to develop new methods based on AI and use new findings from large-scale experiments.”
Revolutionary algorithm
This study could take years if it were not for the rapid developments we have seen in the AI field. In 2020, DeepMind, a British company, presented the AlphaFold algorithm, which can accurately predict individual proteins' structure without homology to known protein structures.
In summer 2021, the program was made available to the research community when the source code was released. Elofsson recalls:
“I remember I was at home on our jetty when I suddenly saw a whole load of posts on Twitter by researchers who were as excited as I was.”
By the end of 2022, several hundred scientific articles had been published based on the new technology.
“AlphaFold has completely revolutionized how we study protein structures,” Elofsson enthuses.
AlphaFold is based on “deep learning.” The program has been fed with training data from all known proteins. Other valuable information is also included, such as how proteins interact.
Elofsson and his colleagues quickly demonstrated that the algorithm is a valuable tool for understanding how proteins interact with each other. They examined 65,000 known protein interactions and also managed to model the structure of several thousand of them.
Interdisciplinary research
These positive experiences have led to the current project. It involves researchers at Stockholm University, KTH Royal Institute of Technology and Uppsala University. Participants possess expertise in fields such as bioinformatics, computer science, graph neural networks and mass spectrometry.
The researchers plan to use new machine-learning methods to identify different proteins, known as protoforms. They can then go on to study how they interact with each other using AI technology, among other things.
Their findings can then be verified in various ways. Cross-linking is used to study which proteins bind to specific DNA sequences. Mass spectrometry is a fundamental means of determining the composition of protein complexes and is a sensitive and speedy technology that detects proteins even in low concentrations.
A biological milestone
The project faces international competition, but Elofsson believes Sweden is up there with the best. A key factor is the billion kronor investment by Knut and Alice Wallenberg Foundation in data-driven life sciences and Berzelius, Sweden’s fastest supercomputer for AI and machine learning.
“Biology is becoming increasingly data-driven, and it is essential to develop methods that can be used by everyone in order to drive research forward. So it is gratifying that so much of the project contributes to methodological development,” says Elofsson.
The research will likely provide a complete picture of the molecular components of a human cell and their interactions. This would represent a milestone for biology and add to our knowledge of complex biological processes and diseases at the molecular level.
“We would be able to perform simulations of entire cells and study their functions at a level of detail that was previously inconceivable,” Elofsson concludes.
Text Nils Johan Tjärnlund
Translation Maxwell Arding
Photo Magnus Bergström