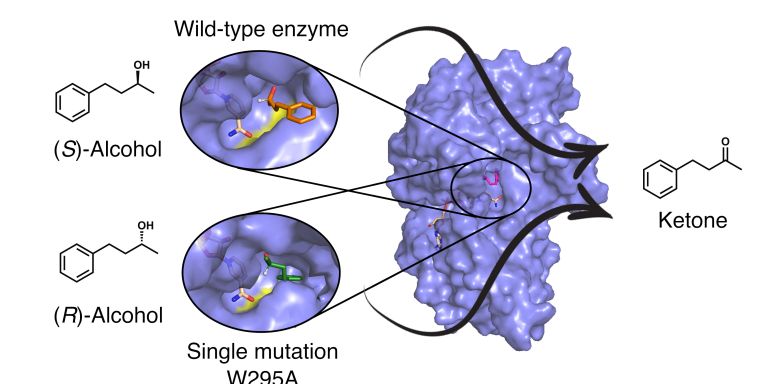
Lynn Kamerlin is using supercomputers to learn how proteins have developed over billions of years of evolution. Most of all, she is interested in enzymes. She is using advanced biomolecular simulations to design new effective enzymes. One strategy is by further developing the catalytic abilities of ancient proteins.
Lynn Kamerlin
Professor of Cell and Molecular Biology
Wallenberg Academy Fellow/Wallenberg Scholar
Institution:
Uppsala University
Research field:
Computational biological studies of enzyme evolution and design
Kamerlin explains that without enzymes biological life would be impossible. Enzymes are the proteins that catalyze chemical reactions in cells. They can increase the rate of reaction several million times over.
“We are alive because chemical reactions constantly take place in the body. When they stop – life stops. So understanding where enzymes come from and how their functions have evolved is to understand the evolution of life down to the tiniest molecule.”
Kamerlin, who is a professor of cell and molecular biology at Uppsala University, likens the life process to a large factory in which many parts communicate with each other and work together – and where the timing has to be just right if everything is to work as it should.
“It’s incredibly complicated. You can learn a lot about these biological processes in a normal wet lab, and build fairly simple models. My research team is trying to contribute more complex models. In our virtual lab we can use chemistry, mathematics and computer science to explain the underlying processes controlling life. And when we have understood it, we may be able to develop new drugs to treat diseases or innovations for a sustainable future, for example.”
Breaking down pesticides and plastic
Enzymes can be fabricated synthetically and tailored to acquire specific traits. They are important catalytic tools in industrial processes, such as those used to manufacture drugs and chemicals. The ability to make these synthetic enzymes requires knowledge of how nature’s own proteins work.
Kamerlin is using advanced computational models and powerful computer clusters to study the evolution of an enzyme that breaks down a toxic substance in nerve gases and pesticides.
“In nature there are enzymes that have evolved over just a few decades to be able to break down organophosphates – completely unnatural compounds that are found in nerve gases and pesticides. In some parts of the world organophosphates are still used in agriculture, even though they’re highly toxic. If we can learn from nature’s own degrading enzymes, and modify them, we’ll hopefully be able to contribute to the development of a cure for organophosphate poisoning.”
The research team is also studying enzymes capable of breaking down plastic.
“We have an exciting collaboration with fellow researchers in Chile. They’ve identified new enzymes that can break down plastic in extremely cold conditions, like those in Antarctica. It would be interesting to see whether they can be engineered to make them better at it. We’re also really interested in developing enzymes that can break down pollutants in water.”
Redesigning an ancient enzyme
Thanks to the Wallenberg Scholar grant, Kamerlin can pursue her research in even more directions. The research team has redesigned an enzyme that is several billions of years old from which many generations of new enzymes have been developed. They want to use it to design new enzymes with unique reaction properties.
“Research is about working hard, but also about luck – you have to be in the right place at the right time. Receiving this Scholar grant is wonderful because it enables us to do cool research that wouldn’t have been possible otherwise.”
Kamerlin explains that they use bioinformatic techniques to determine the genome sequence of ancient proteins. Bioinformatics is an interdisciplinary computation method for analyzing biological data.
“When we have identified the DNA sequence, we can recreate the ancient protein. We then compare it with the molecular and atomic structure of modern synthetic enzymes. Our colleagues in Uppsala, elsewhere in Sweden, and around the world, then build the actual enzymes and take measurements of them in the laboratory. We know, for example that at an early stage of evolution proteins were fairly stable when subjected to heat, whereas modern proteins disintegrate if they are heated up.”
In one such case, we have used computational approaches to redesign an ancient protein, with the redesigned enzyme being competitive against the average modern enzymes towards its natural substrate when a specific non-natural catalytic reaction was tested.
“But one of the engineered enzymes was really good. It had been designed using computational chemistry, a purely theoretical model, and directed evolution. So we wanted to see whether we could achieve the same efficiency based on our ancient enzyme. And we did in fact demonstrate that by using a fairly simple design strategy involving bioinformatic techniques and computational design, we could create a non-natural enzyme that could break down a non-natural substrate almost as well as the best current synthetic enzyme after many rounds of optimization by laboratory evolution. That is something we’re proud of.”
Hot research field
In the past decade alone, two Nobel prizes have been awarded for breakthroughs in Kamerlin’s research field. In 2013 her former supervisor Arieh Warshel at the University of Southern California won the Nobel Prize in Chemistry for the development of multiscale models for complex chemical systems. Five years later, in 2018, Francis Arnold at Caltech in the U.S. was awarded the prize for directed evolution and optimization of enzymes. Kamerlin is using the methods of both Nobel prizewinners in her studies of enzyme evolution and design.
“It’s very gratifying of course – it shows that the field is important. It was also a boost for all of us working in the field.”
Text Susanne Rosén
Translation Maxwell Arding
Photo Marina Corbella, Mikael Wallenstedt, Rory Crean, Peter Casson, Magnus Bergström